Parallel Computing on Windows with R
By default, R doesn’t acknowledge that your computer has multiple logical or even physical cores, which will often result in your long repetitive job tying up one core for hours and hours while the rest laze about.
There are several fixes for this. A smooth package for repetitive work is Revolution Analytics’ foreach package, along with a corresponding parallel backend. The most important determinant for which one chooses is platform - OS X and other Unices can make use of the multicore package, however since it relies on the fork() system call and because Windows doesn’t implement this, Windows users must use an alternative like SNOW/doSNOW, which also exists on other platforms. These can be used to control several machines as a cluster, however for the purposes of a single machine a virtual cluster can also be created. That’s what I do here.
First, one initializes:
library(foreach)
library(doSNOW)
cl <- makeCluster(2)
registerDoSNOW(cl)
The cl command creates our cluster object, and registerDoSNOW allows it to be used. Then, given a function foo(), one need only run
outputList <- foreach(i=1:1000) %dopar% foo(args)
(you can do testing on one core by swapping %dopar% for %do%)
One wrinkle in this is that each instance of foo effectively runs in its own environment, and so other libraries needed (e.g. the survey package) need to be loaded within foo().
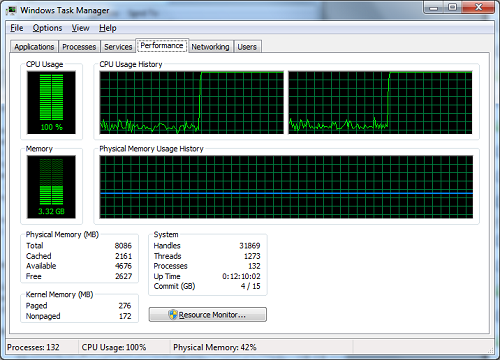
Looks satisfying, doesn’t it?
